A major obstacle in current valve disease research is the lack of high-quality homogeneous functional heart valve cells.Human induced pluripotent stem cell (hiPSCs)-derived heart valve cells may help to address this challenge. However, there are no established protocols for inducing differentiation of hiPSCs into functional heart valve cells, and the networks that mediate differentiation have not been fully elucidated.
On February 15, 2024, a research paper entitled “Directed Differentiation of Human Induced Pluripotent Stem Cells to Heart Valve Cells” was published online by Circulation (IF=38) with the joint communication of Nengguo Dong and Weihua Qiao from Huazhong University of Science and Technology. Valve Cells", which developed an effective protocol to induce differentiation of hiPSCs into functional hiPSCs-derived valve endothelial-like cells and hiPSCs-derived valve mesenchymal-like cells. After 6 d of differentiation and CD144 magnetic bead sorting, approximately 70% CD144+ cells and 30% CD144 - cells were obtained. Based on single-cell RNA sequencing data, CD144+ cells and CD144 -cells were found to be highly similar to primary heart valve endothelial cells and primary heart valve mesenchymal cells in terms of gene expression profiles. In addition, CD144+ cells had functions typical of primary cardiac valve endothelial cells, including tube formation, uptake of low-density lipoprotein, endothelial nitric oxide synthase production, and response to shear stress.
Meanwhile, CD144 -cells secrete collagen and matrix metalloproteinases and differentiate into osteoblastic or lipogenic cell lines, such as primary heart valve mesenchymal cells. Therefore, this study identified CD144+ cells and CD144 - cells as hiPSCs-derived valve endothelial-like cells and hiPSCs-derived valve mesenchymal-like cells, respectively. The trajectory of heart valve cell differentiation was demonstrated to be consistent with embryonic valve development by single-cell RNA sequencing analysis. The major switch genes (NOTCH1, HEY1, and MEF2C), signaling pathways (TGF-β, Wnt, and NOTCH), and transcription factors (MSX1, SP5, and MECOM) mediating differentiation were also identified. Finally, it was found that hiPSCs-derived valvular mesenchymal-like cells may be derived from hiPSCs-derived valvular endothelial-like cells, which undergo endocardial mesenchymal transformation. This study reports, for the first time, an effective strategy for generating functional hiPSCs-derived valve endothelial-like cells and hiPSCs-derived valve mesenchymal-like cells from hiPSCs, and elucidates the differentiation trajectory and transcriptional dynamics of hiPSCs differentiating into cardiac valve cells.

Valvular heart disease (VHD) is an important and difficult-to-treat cardiovascular disease that affects human health, including rheumatic, calcific, infectious, and congenital VHD.Recent epidemiologic studies have shown that VHD affects approximately 74 million people worldwide each year, with more than 539,000 deaths. However, there are no effective therapeutic agents for VHD because the pathogenesis of the disease is unknown. In conclusion, normal heart valves are mainly composed of two types of cells, valve endothelial cells (VECs) and valve interstitial cells (VICs), which play a crucial role in the pathogenesis of VHD. Currently, in vitro studies of VHD mainly rely on primary VECs and VICs.However, primary VECs and VICs are difficult to obtain and they have low proliferative capacity and high heterogeneity, which is a research advancement in the use of VHD for drug development. Therefore, it is necessary to explore other alternative methods to generate a continuous source of homogenous human VECs and VICs.
The recent emergence of techniques for inducing the differentiation of human pluripotent stem cells (hiPSCs) into a variety of disease-associated cell types with increasing accuracy provides an unprecedented opportunity to generate large numbers of homogeneous human patient-specific cells. hiPSCs can now be efficiently reprogrammed into disease-associated cell types, including hipsc-derived cardiomyocytes and hiPSCs-derived neurons. However, there are no well-established protocols for inducing differentiation of hiPSCs into functional VECs and VICs. several previous studies have reported the possibility of generating endocardial/valvular endothelial-like cells from hiPSCs, which express a number of markers of the endocardial spectrum. nachlas et al. have demonstrated that iPSCs-derived MSCs can mature in hydrogels into VICs-like cells. Although the results of these studies are promising, none of them reported a single protocol for the co-generation of functional VECs and VICs from hiPSCs. therefore, there is still a need to study and develop new protocols to generate hiPSCs-derived valve endothelial-like cells (hiVECs) and hiPSCs-derived valve mesenchymal-like cells with functions similar to those of progenitor valve cells ( hiVICs).
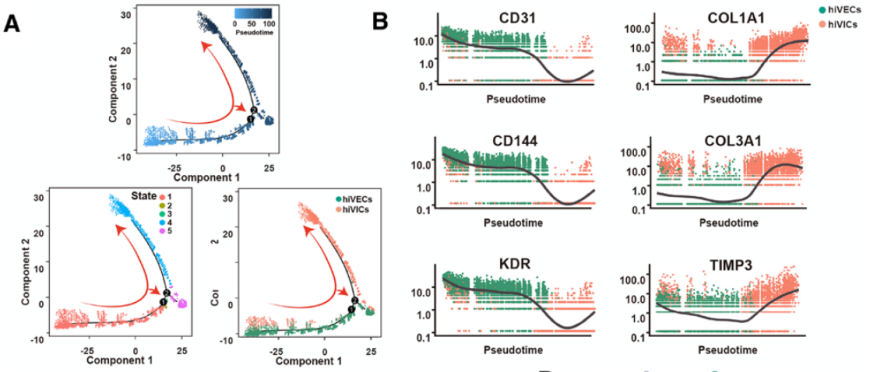
Pseudotemporal trajectories of hiVECs and hiVICs (figure derived from Circulation )
Differentiation of stem cells into desired cell lines requires alternate developmental pathways and often results in unwanted cell types. Although previous studies reported the expression of some marker genes during valve endothelial-like cell differentiation, the exact mechanism by which heart valve cells differentiate from hiPSCs is unknown. Therefore, it is necessary to comprehensively characterize the transcriptional dynamics throughout the differentiation process. Recent advances in single-cell RNA sequencing (scRNAseq) provide an excellent opportunity to comprehensively track transcriptional dynamics throughout differentiation at single-cell resolution.
This study developed, for the first time, a stable and efficient protocol for generating functional hiVECs and hiVICs from hiPSCs. hiVECs and hiVICs are highly similar to primary valve cells in terms of gene expression profile and function. In addition, both cells exhibited a greater proliferative capacity than primary heart valve cells. This protocol provides an unprecedented access to high-quality homogeneous functional VECs and VICs, which can be used to establish toxicology platforms and VHD models and construct tissue-engineered heart valves. In addition, longitudinal scRNA-seq was performed to reveal differentiation trajectories and transcriptional dynamics during the differentiation of hiPSCs to heart valve cells. These findings contribute to further understanding of the mechanisms of embryonic heart valve differentiation and provide new candidates for studying embryonic heart valve development. development of hiPSCs-derived VHD models and heart valve substitutes will contribute to improved therapeutic approaches for VHD.


